Neo4j GraphRAG Python 包
Neo4j GraphRAG 包是一个全面的 Python 库,用于构建生成式 AI 应用程序。它通过一个管道支持知识图谱创建,该管道从非结构化文本中提取实体、生成嵌入,并在 Neo4j 中创建图谱。该包还提供了多种检索器,用于图谱搜索、向量搜索以及与向量数据库的集成。
用法 - 生物医学知识图谱示例
使用 SimpleKGPipeline 进行首次知识图谱构建
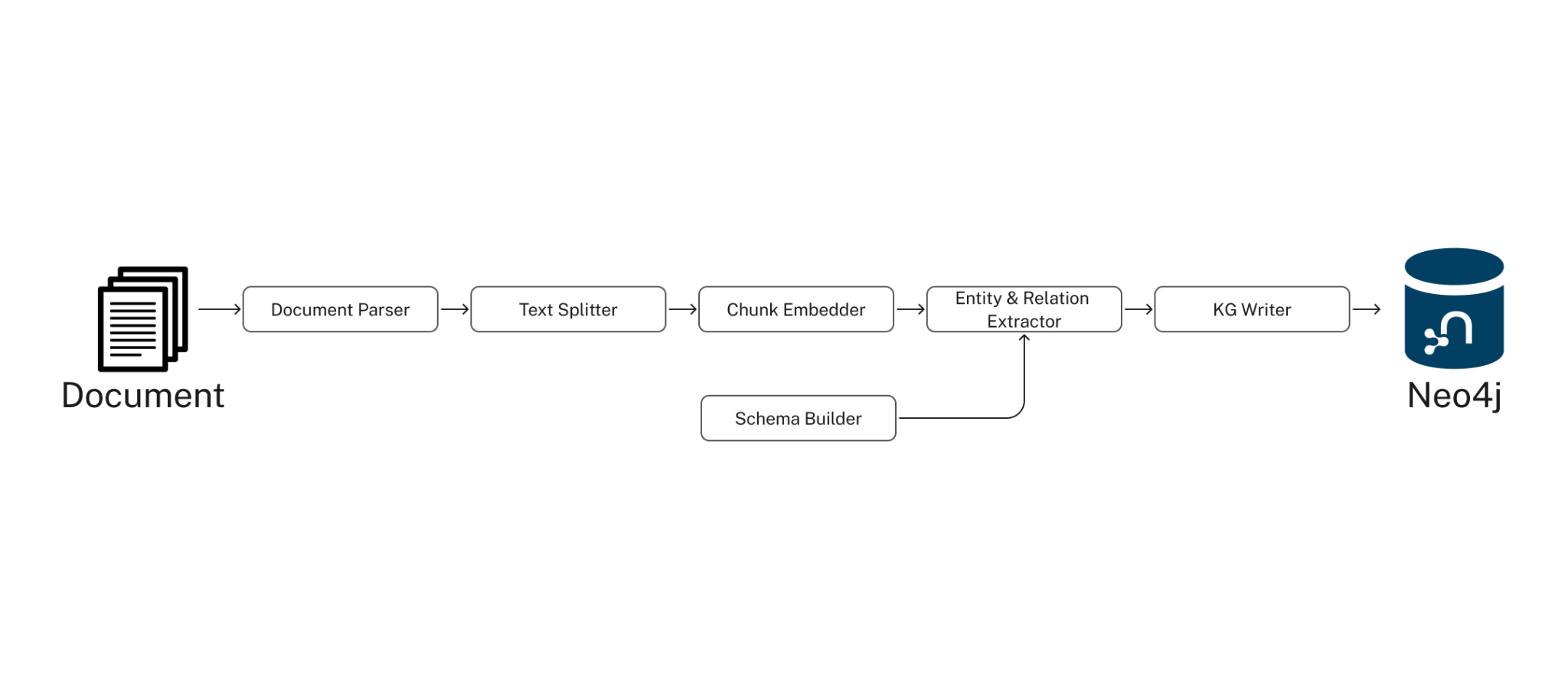
设置 Neo4j 连接、模式和基础模型(LLM、嵌入)以及提取提示模板。
# Neo4j Driver
import neo4j
neo4j_driver = neo4j.GraphDatabase.driver(NEO4J_URI,
auth=(NEO4J_USERNAME, NEO4J_PASSWORD))
# LLM and Embedding Model
from neo4j_graphrag.llm import OpenAILLM
from neo4j_graphrag.embeddings.openai import OpenAIEmbeddings
llm=OpenAILLM(
model_name="gpt-4o-mini",
model_params={
"response_format": {"type": "json_object"}, # use json_object formatting for best results
"temperature": 0 # turning temperature down for more deterministic results
}
)
# Graph Schema Setup
basic_node_labels = ["Object", "Entity", "Group", "Person", "Organization", "Place"]
academic_node_labels = ["ArticleOrPaper", "PublicationOrJournal"]
medical_node_labels = ["Anatomy", "BiologicalProcess", "Cell", "CellularComponent",
"CellType", "Condition", "Disease", "Drug",
"EffectOrPhenotype", "Exposure", "GeneOrProtein", "Molecule",
"MolecularFunction", "Pathway"]
node_labels = basic_node_labels + academic_node_labels + medical_node_labels
# define relationship types
rel_types = ["ACTIVATES", "AFFECTS", "ASSESSES", "ASSOCIATED_WITH", "AUTHORED",
"BIOMARKER_FOR", …]
#create text embedder
embedder = OpenAIEmbeddings()
# define prompt template
prompt_template = '''
You are a medical researcher tasks with extracting information from papers
and structuring it in a property graph to inform further medical and research Q&A.
Extract the entities (nodes) and specify their type from the following Input text.
Also extract the relationships between these nodes. the relationship direction goes from the start node to the end node.
Return result as JSON using the following format:
{{"nodes": [ {{"id": "0", "label": "the type of entity", "properties": {{"name": "name of entity" }} }}],
"relationships": [{{"type": "TYPE_OF_RELATIONSHIP", "start_node_id": "0", "end_node_id": "1", "properties": {{"details": "Description of the relationship"}} }}] }}
...
Use only fhe following nodes and relationships:
{schema}
Assign a unique ID (string) to each node, and reuse it to define relationships.
Do respect the source and target node types for relationship and the relationship direction.
Do not return any additional information other than the JSON in it.
Examples:
{examples}
Input text:
{text}
'''知识图谱管道设置与执行(含示例 PDF)
# Knowledge Graph Builder
from neo4j_graphrag.experimental.components.text_splitters.fixed_size_splitter import FixedSizeSplitter
from neo4j_graphrag.experimental.pipeline.kg_builder import SimpleKGPipeline
kg_builder_pdf = SimpleKGPipeline(
llm=ex_llm,
driver=driver,
text_splitter=FixedSizeSplitter(chunk_size=500, chunk_overlap=100),
embedder=embedder,
entities=node_labels,
relations=rel_types,
prompt_template=prompt_template,
from_pdf=True
)
pdf_file_paths = ['truncated-pdfs/biomolecules-11-00928-v2-trunc.pdf',
'truncated-pdfs/GAP-between-patients-and-clinicians_2023_Best-Practice-trunc.pdf',
'truncated-pdfs/pgpm-13-39-trunc.pdf']
for path in pdf_file_paths:
print(f"Processing : {path}")
pdf_result = await kg_builder_pdf.run_async(file_path=path)
print(f"Result: {pdf_result}")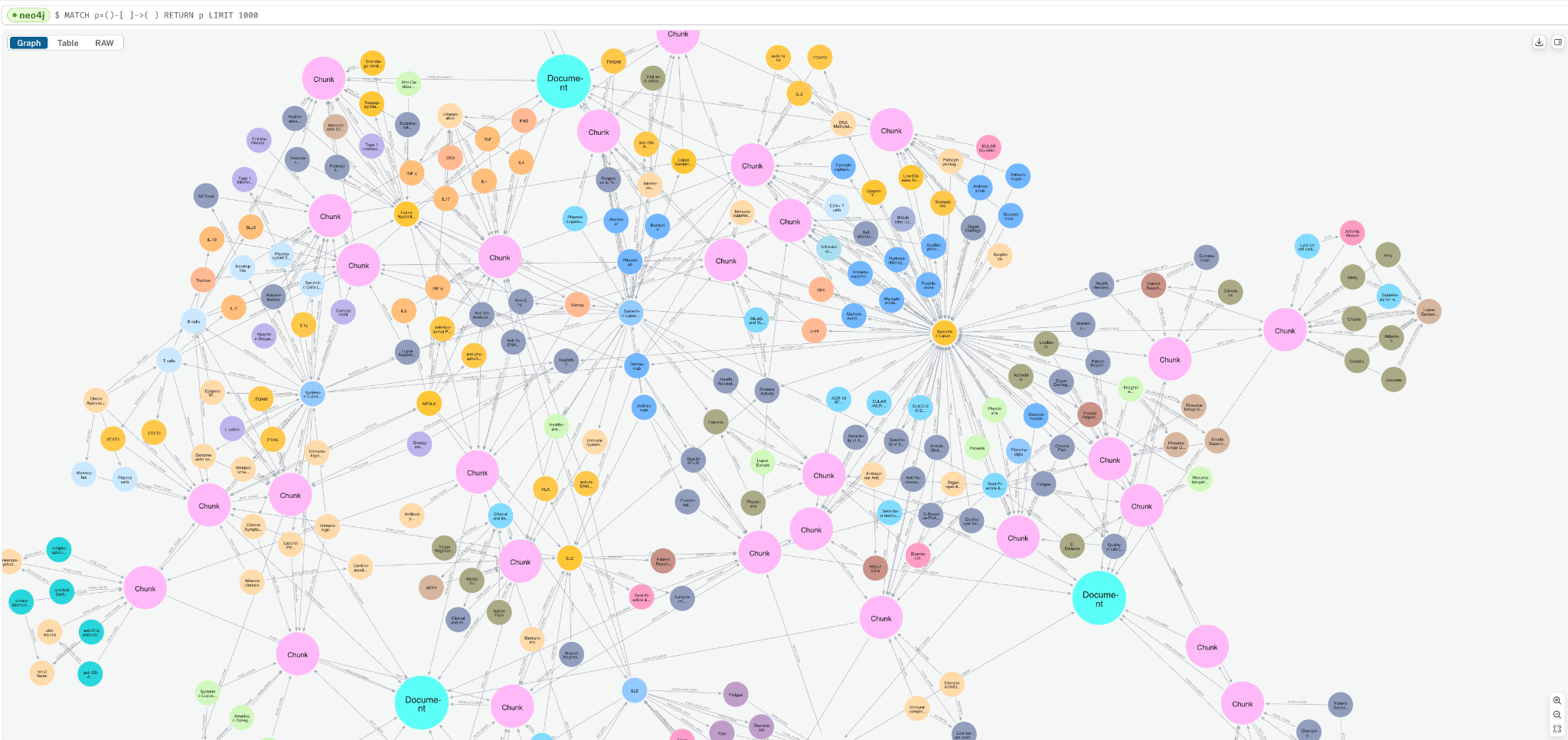
然后使用 VectorCypher 检索器运行 GraphRAG 搜索。
from neo4j_graphrag.indexes import create_vector_index
create_vector_index(driver, name="text_embeddings", label="Chunk",
embedding_property="embedding", dimensions=1536, similarity_fn="cosine")
# Vector Retriever
from neo4j_graphrag.retrievers import VectorRetriever
vector_retriever = VectorRetriever(
driver,
index_name="text_embeddings",
embedder=embedder,
return_properties=["text"],
)
# GraphRAG Vector Cypher Retriever
from neo4j_graphrag.retrievers import VectorCypherRetriever
graph_retriever = VectorCypherRetriever(
driver,
index_name="text_embeddings",
embedder=embedder,
retrieval_query="""
//1) Go out 2-3 hops in the entity graph and get relationships
WITH node AS chunk
MATCH (chunk)<-[:FROM_CHUNK]-(entity)-[relList:!FROM_CHUNK]-{1,2}(nb)
UNWIND relList AS rel
//2) collect relationships and text chunks
WITH collect(DISTINCT chunk) AS chunks, collect(DISTINCT rel) AS rels
//3) format and return context
RETURN apoc.text.join([c in chunks | c.text], '\n') +
apoc.text.join([r in rels |
startNode(r).name+' - '+type(r)+' '+r.details+' -> '+endNode(r).name],
'\n') AS info
"""
)
llm = LLM(model_name="gpt-4o", model_params={"temperature": 0.0})
rag_template = RagTemplate(template='''Answer the Question using the following Context. Only respond with information mentioned in the Context. Do not inject any speculative information not mentioned.
# Question:
{query_text}
# Context:
{context}
# Answer:
''', expected_inputs=['query_text', 'context'])
vector_rag = GraphRAG(llm=llm, retriever=vector_retriever, prompt_template=rag_template)
graph_rag = GraphRAG(llm=llm, retriever=graph_retriever, prompt_template=rag_template)
q = "Can you summarize systemic lupus erythematosus (SLE)? including common effects, biomarkers, and treatments? Provide in detailed list format."
vector_rag.search(q, retriever_config={'top_k':5}).answer
graph_rag.search(q, retriever_config={'top_k':5}).answer
